

We are intrigued by how proteins work. The genetic code determines the amino acid sequence of proteins, which in turn determines their 3D-structure. The precise 3D-arrangement of different types of atoms inside proteins allows them to perform the many types of functions that are needed to keep the cell alive. Therefore, visualising the 3D-structures of proteins is a powerful way to find out more about how they work. Research in our lab focuses on different aspects of protein structure determination.
One line of research is the development of image processing methods to determine protein structures from electron cryo-microscopy (cryo-EM) images. We implement these methods in our open-source computer program RELION. This program is used to solve protein structures in many cryo-EM labs worldwide.
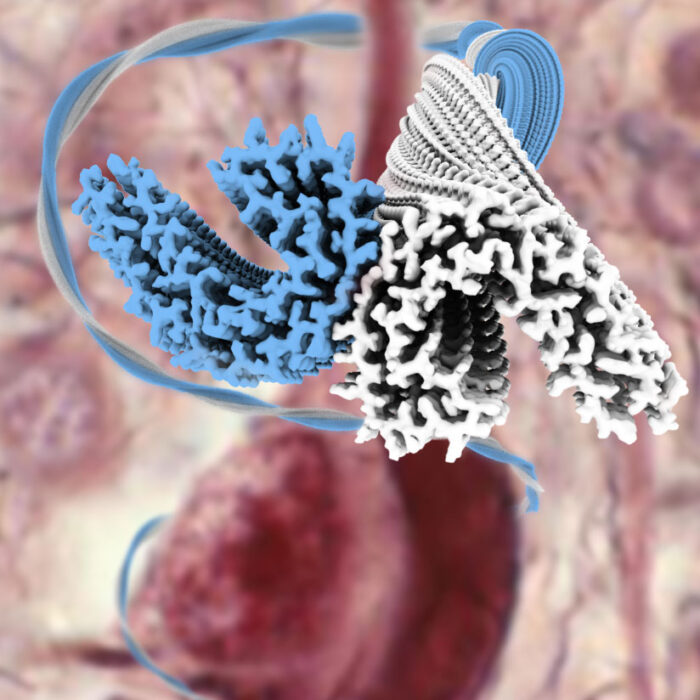
In another line of research, we use experimental cryo-EM together with our own image processing developments to discover new protein structures. Instead of solving structures of proteins in their healthy, functional state we focus on protein structures that are associated with disease. For this purpose, we extract amyloid filaments from the brains of individuals with different types of neurodegenerative diseases, like amyloid filaments made of the protein Tau from Alzheimer’s disease (image above) or chronic traumatic encephalopathy. This work has revealed that the same protein may adopt surprisingly different structures in different diseases, and ongoing efforts are aimed at understanding how this happens. This line of research is done in close collaboration with the Goedert group.
Selected Papers
- B. Falcon, J. Zivanov, W. Zhang, A.G. Murzin, H.J. Garringer, R. Vidal, R.A. Crowther, K.L. Newell, B. Ghetti, M. Goedert & S.H.W. Scheres (2019)
Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules
Nature 568: 420-423. - J. Zivanov, T. Nakane, B. Forsberg, D. Kimanius, W.J.H. Hagen, E. Lindahl & S.H.W. Scheres (2018)
RELION-3: new tools for automated high-resolution cryo-EM structure determination
eLife 7: e42166. - A.W.P. Fitzpatrick, B. Falcon, S. He, A.G. Murzin, G. Murshudov, H.J. Garringer, R.A. Crowther, B. Ghetti, M. Goedert & S.H.W. Scheres (2017)
Cryo-EM structures of tau filaments from Alzheimer's disease
Nature 547: 185-346. - S.H.W. Scheres (2012)
RELION: Implementation of a Bayesian approach to cryo-EM structure determination
J. Struc. Biol. 180: 519-530.
Group Members
- Chloe Fisher
- Kiarash Jamali
- Simon Knoblauch
- Sofia Lövestam
- Ben Luisi
- Alexey Murzin
- Yiming Qin
- Bogdan Toader
- Kai Yi
- Dongqi Zhang